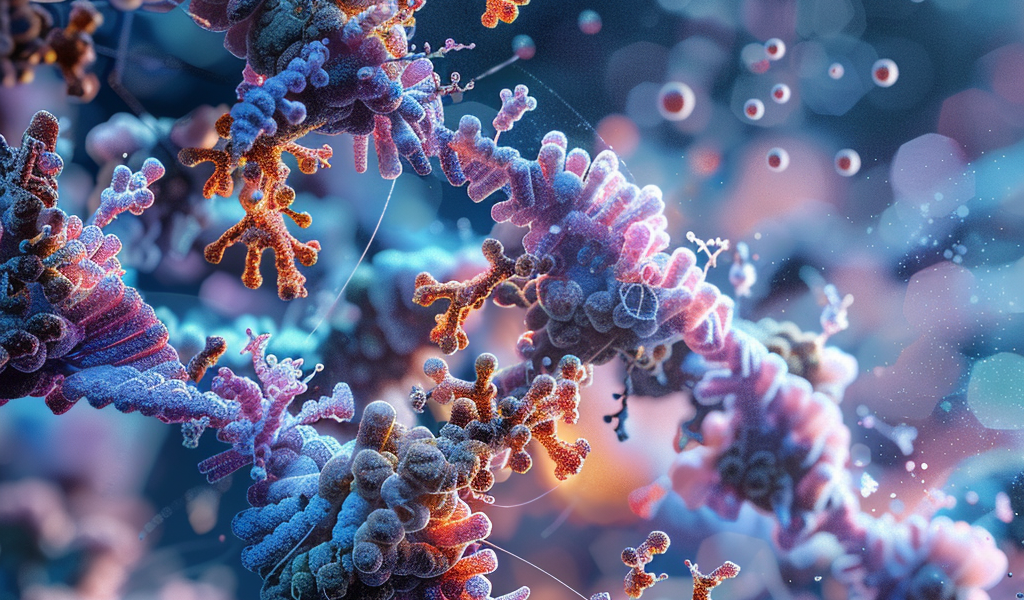
Google DeepMind and Isomorphic Labs have jointly introduced AlphaFold 3, an advanced AI model that aims to revolutionize the understanding of life’s molecules. This cutting-edge technology accurately predicts the structure and interactions of essential components such as proteins, DNA, RNA, and ligands, offering unprecedented insights into the biological world and drug discovery.
Within the intricate framework of every living organism, molecular machines comprising proteins, DNA, and various molecules operate collectively to drive life’s processes. AlphaFold 3, as detailed in a recent publication in Nature, represents a groundbreaking development in predictive modeling, showcasing remarkable accuracy in forecasting the interactions of diverse molecules. Notably, the model demonstrates a significant improvement of up to 50% in predicting protein interactions compared to existing methods, with a doubling of accuracy in crucial interaction categories.
The potential impact of AlphaFold 3 extends beyond theoretical advancements, with a focus on practical applications in drug design and pharmaceutical research. The newly launched AlphaFold Server provides scientists with access to a range of capabilities at no cost, offering a user-friendly platform for innovative research endeavors. Collaborations between Isomorphic Labs and pharmaceutical companies are already underway, leveraging AlphaFold 3 to address real-world challenges in drug development and enhance patient care through the creation of novel treatments.
Building upon the success of its predecessor, AlphaFold 2, which made significant strides in protein structure prediction, the latest iteration expands its scope to encompass a broader array of biomolecules. This expansion opens up avenues for transformative scientific discoveries, including advancements in biorenewable materials, crop resilience, drug design, and genomics research.
One notable application of AlphaFold 3 is its structural prediction of the spike protein of a common cold virus (Coronavirus OC43), demonstrating a high level of accuracy in depicting the protein’s interactions with antibodies and simple sugars. This detailed insight into immune-system processes not only enhances our understanding of coronaviruses, including COVID-19, but also paves the way for improved treatment strategies.
By harnessing the predictive capabilities of AlphaFold 3, researchers are poised to delve deeper into the complexities of molecular interactions, unlocking new possibilities for scientific exploration and innovation in the realm of life sciences.